Personalized Medicine Research
Precision Oncology
Precision oncology is a treatment strategy in cancer medicine that establishes
Clinical Variant Annotation Pipeline (ClinVAP)
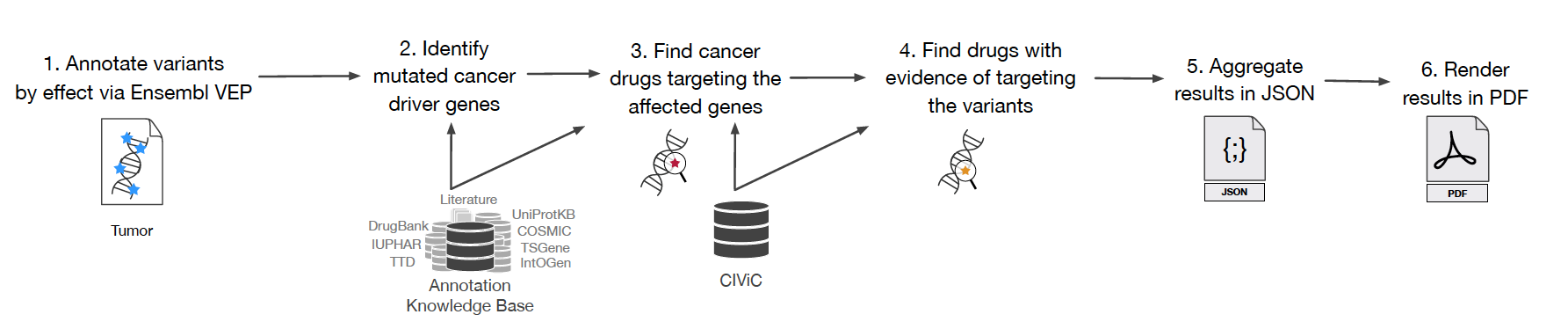
Clinical application of precision oncology necessitates the employment of fast, robust and automated pipelines to process genomics data and retrieve the mutational profile of the patients. To meet this need, we developed ClinVAP which translates the long list of variants to
MyDrug
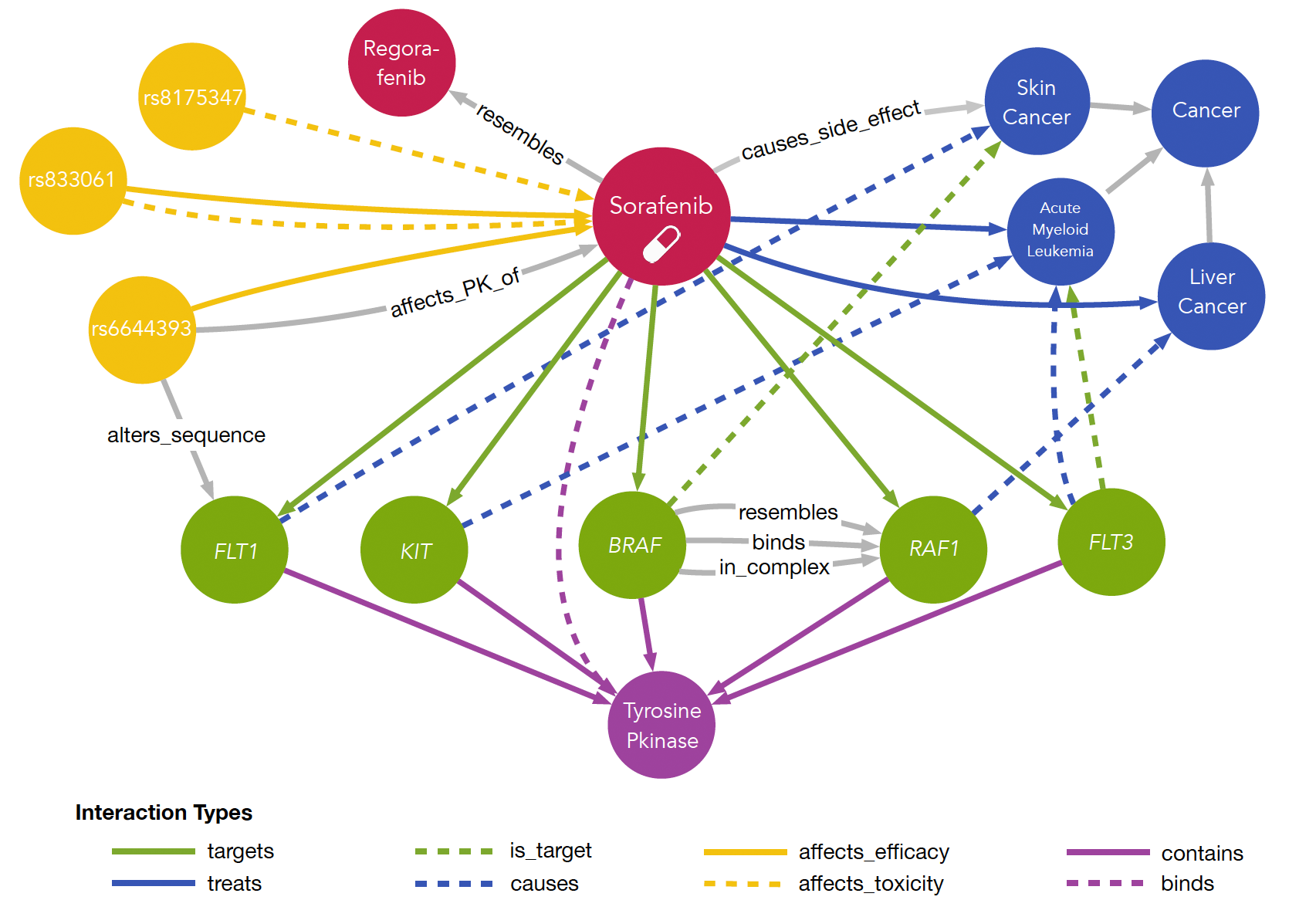
The impact of the variants on the patient’s response to therapeutics makes the establishment of the tailored treatment strategies based on the patient’s genetic profile of crucial importance. For personalized treatments, the factors affecting the patient’s response to drugs needs to be uncovered by examining the mechanisms of drug efficacy and resistance, and by assigning new indications to existing drugs based on cellular interaction networks. Within this scope, we created MyDrug, a heterogeneous information network collating information from various curated resources about the dynamics between therapeutics and the molecular disease mechanisms along with the clinical evidence on drug efficacy.
Network Biology
The cellular machinery is comprised of many individual metabolic, regulatory and signalling events. Put together they build vast networks of interconnected reactions, interactions and relations. Our group researches various methods to leverage the information in these networks as well as to analyze clinical data in that context for additional insight.
DeRegNet
We develop DeRegNet, a software to find deregulated subnetworks in large directed biological networks based on scores that were derived from various omics data. The underlying method relies on fractional integer programming to arrive at optimal solutions. This open-source software allows many flexible use cases for functional annotation and hypothesis-generation for omics based research.
Biological Networks in Graph Databases
To utilize the analytical power of graph databases we are developing an application to persist biological models available in the SBML format (http://sbml.org/Main_Page) in graph databases. The resulting graph representations allow the creation of network mappings (e.g. regulatory networks, metabolic networks
Subnetwork-Guided Analyses
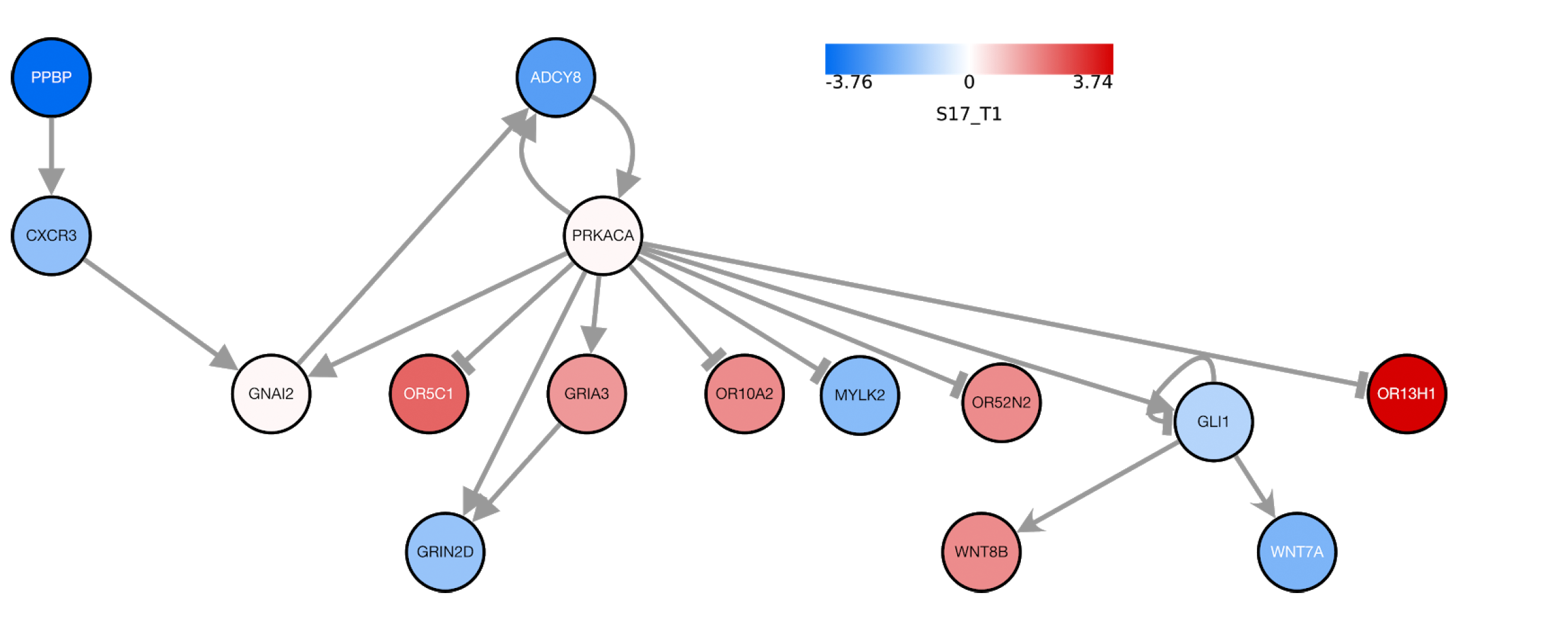
By making use of the integrated systems biology knowledge, the deregulated subnetworks generated by DeRegNet allow us to explore putative disease mechanisms and relations that go beyond already disseminated pathway knowledge. Thus, one of our current focal points is to assess how to best utilize these subnetworks for Machine Learning applications in personalized medicine, such as disease prediction or patient stratification.
Visual Analytics
To prevent, diagnose and treat disease, personalized medicine uses information about a person’s genes, proteins, and environment. Instead of poring over spreadsheets or reports, it is easier to use charts and graphs to visualize these large amounts of data as the human brain is trained to see and understand trends, outliers, and patterns in data visually. Therefore, we are working on new ways to visually include patient data to the MTBs. Our first step is to integrate an interactive visualization of the DeRegNets into the MTB to show genes that are possibly related to